Research
I currently combine working as a consultant in machine learning with working as an independent researcher with a focus on open science. You can follow my open research journey here:
I previously worked as a researcher in academia for over 12 years. My research took me into diverse fields of study including marine mammal biology, computational biology, protein folding, artificial life, complex systems analysis, plant biology, genomics, marine molecular developmental biology and RNA biology.
Read on to learn about my earlier research interests in ecogenomics and the fascinating genome of the zooplankton, Oikopleura dioica.
You can also read about my PhD research in the fields of complex systems and artificial life.
Developmental Ecogenomics
Genome-environment interactions
A complex multicellular organism develops from an egg by following the instruction manual from its parents that it keeps in its genome. The information is all there waiting to be read by the molecular machinery of the egg and the DNA manual is copied into every cell. But once development gets underway, the same instructions are not read by every cell. The genome is subject to constant re-wiring — muting some parts and activating others — to bring about critical transitions at specific times and in specific places.
Back in academia, my main research interest was in trying to understand how the genome is rewired to orchestrate developmental transitions and how this rewiring responds to changes in the environment and determines alternate life history strategies.

Oikopleura dioica
This zooplankton, living in oceans across the globe, is among the closest living relatives to vertebrates. It has a small, shuffled genome and it builds complex filter-feeding houses that it sheds several times a day – making marine snow and capturing carbon in the process.
Oikopleura is a fascinating animal for studying how the genome is rewired during development.
I spent 8 years working on it as a postdoctoral researcher at the CBU and Sars Centre in Bergen, Norway.
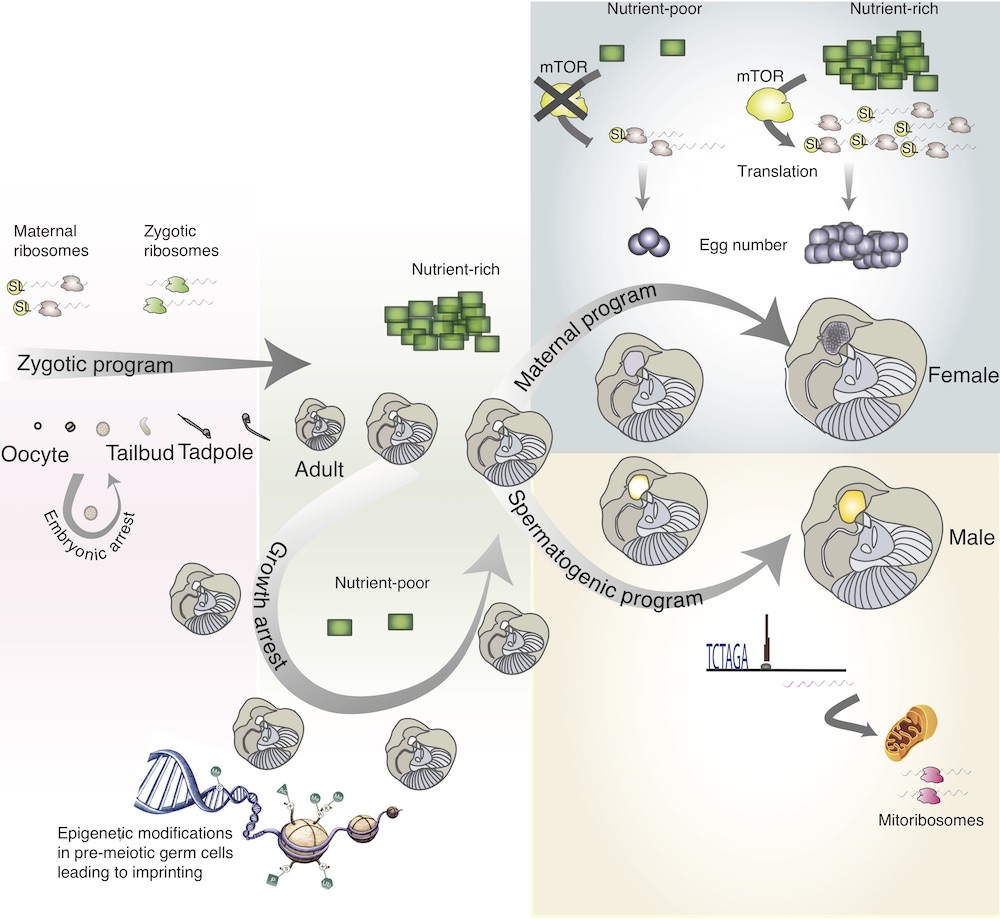
You can see a summary of my areas of research in Oikopleura in the figure below. Read on for the details.

Research
RNA trans-splicing as a regulator of egg numbers
My favourite topic!
Oikopleura has a very special way of processing many of the messenger RNA molecules it transcribes from its genes: it adds an extra sequence to one end. This is called RNA trans-splicing. Amazingly, despite several decades of research into the few animals that do this, the function of the extra sequence was largely mysterious.
I developed and tested the hypothesis that RNA trans-splicing works as a way of regulating the egg numbers in Oikopleura according to how much food the zooplankton has in its environment via an ancient molecular regulator of growth. This regulation might be helping the population respond to algal blooms in the ocean.
This goes beyond the population levels of Oikopleura: it’s a fascinating example of how the molecular blueprint of life can receive cues from the wider environment.
You can read more about this in three of my publications on the topic: the first one shows that most of the genes that have this extra tag are maternal (i.e. stocked in eggs); the second is a commentary that fleshes out the hypothesis in more detail; the third puts the hypothesis to the test.
Research

Environmental cues for exiting growth arrest
In the lab, if you culture Oikopleura at high enough density and restrict their food they will arrest their development right before they start their reproductive program. They stay in stasis until conditions are better for growth. I studied what environmental triggers cause them to leave stasis, how this is controlled at the level of the genome and how it is related to RNA trans-splicing.
You can read more about this in two of my papers on trans-splicing here and here.
Research
Promoter codes for developmental transitions
When the cellular machinery has to read the genome it needs to know what parts to read and where to start. Promoters are landing sites in the DNA for this machinery and they are written into the DNA using special codes that mark what genes should be switched on and when.
I studied these codes in Oikopleura and used machine learning to classify them. You can read more about how in this paper.

Research

Chromatin states
DNA is packaged in the cell using proteins called histones. Histones also have a code that tells the cellular machinery what parts of the DNA should be opened up and read and which parts should be closed off and silenced. I worked on deciphering this code and how it differs in male and female Oikopleura. You can read more details here.
Research
Genome evolution and house building
One of the most fascinating things about Oikopleura is that it builds this beautifully complex ‘house’ around itself for filter-feeding algae from seawater. It is the possession of this ‘house’ that gave Oikopleura its name (oiko comes from the greek word for house). Every few hours, Oikopleura sheds its house and builds a new one. The discarded houses sink through the ocean as marine snow and trap carbon in the process.
Oikopleura builds its houses using special cells around its body that, together, secrete a particular pattern of molecules that create the different chambers and funnels of the house.
Some of these molecules, called oikosins, are not found in other groups of animals. You can read more about them in this paper.
Complex systems and artificial life
Research
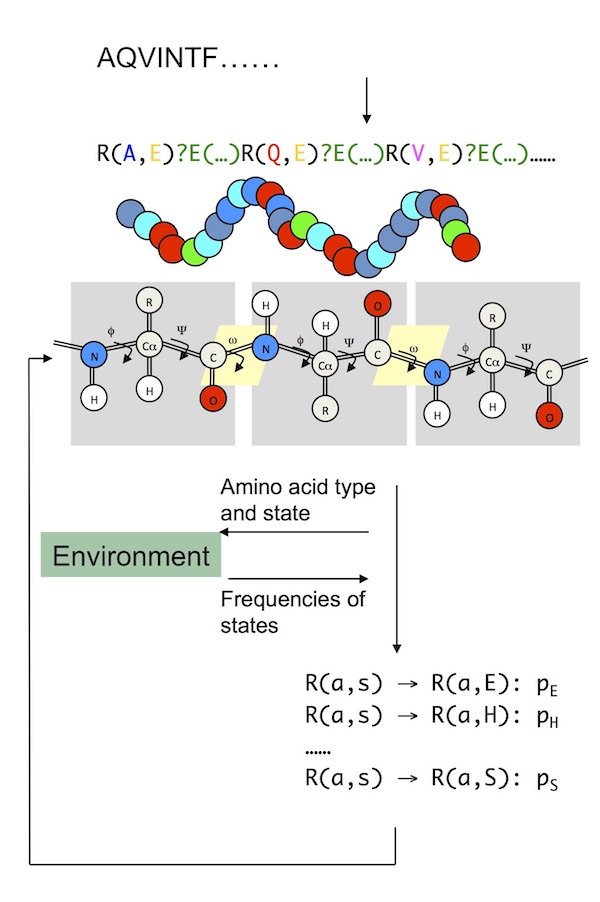
Protein folding with L-systems
The main work horses of the cell are protein molecules. It’s the proteins that bring about development and sustain life. The instructions for building them are encoded in DNA and a large proportion of the genome is devoted to making sure the right proteins are made at the right time and at the right level.
Proteins come in a wide array of shapes and sizes that determine what jobs they do. Some need pockets to help chemical reactions proceed. Others need to form sheets or tubules. Their shape is determined by the sequence of their amino acid building blocks. Yes – another code!
I spent three years studying this particular code during my PhD at the University of York.
I used L-systems, which is a system of mathematics originally used to model plant growth, to explore how simple rewriting rules, together with modelling the protein’s environment, can be used to fold proteins to their final shape. You can read more about this work in three papers I published on the topic here, here and here.